 The Taverna services for Systems Biology (tav4sb) project provides a set of new Web service operations, which extend the functionality of the Taverna Workbench in a domain of systems biology.
The Taverna services for Systems Biology (tav4sb) project provides a set of new Web service operations, which extend the functionality of the Taverna Workbench in a domain of systems biology.
The Taverna Workbench is a tool which facilitates the design and execution of in silico experiments. The experiments are constructed as workflows which can be stored and executed when needed. The building blocks of a workflow are services, also known as processors. Technically, workflow is a set of processors, together with connections between their inputs and outputs. The remote processors are implemented as Web service (WS) operations. Scattered physically throughout computational resources of numerous scientific facilities and combined together, the WSs operations enable a highly complex analysis, surpassing limits of a common workstation.
Operations provided by tav4sb allow for:
- Numerical simulations for the deterministic formulation of a biochemical network model, using the SBML ODE Solver library (SOSlib).
- Probabilistic model checking of Continuous Stochastic Logic (CSL) formula over a CTMC, using PRISM.
- Visualization of data series, such as ODEs trajectories or values of parametrized CSL properties, and probabilistic distribution sampling, using Mathematica.
Service supports the popular Systems Biology Markup Language (SBML), an XML-based data format, to represent kinetic models of biological systems. tav4sb operations are executed in a simple grid environment, integrating heterogeneous software such as Mathematica, PRISM and SBML ODE Solver.
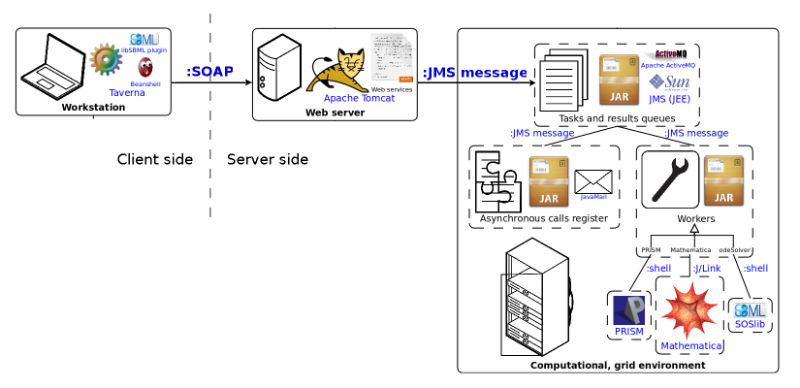
Names of a particular software, technology or standard are written in blue. The communication type is specified on edges which connect components of the system.
Web services utilization flow :
- User defines and runs workflows which include tav4sb Web services.
- Taverna client communicates with Web server via Web services.
- Web server sends computational tasks to the grid environment.
- Grid environment runs computational tasks.
- Depending on service, response might be synchronous or asynchronous (sent via email).
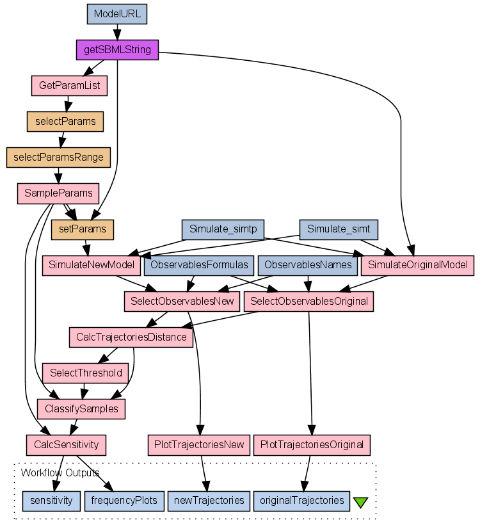
On top of functionality, tav4sb enables the construction of high-level experiments by creating workflows which use provided methods. As an illustration of possibilities offered tav4sb is the multi-parameter sensitivity analysis of biological models, with error calculation via either numerical simulations or the probabilistic model checking technique. MPSA is a Monte Carlo filtering method of sensitivity analysis which maps parameters space into acceptable and unacceptable output regions.
The method works as follows:
- Select parameters to assess.
- Set parameters range.
- Generate independent samples.
- Calculate samples errors.
- Classify samples as acceptable or unacceptable.
- Statistically compare samples sets.
One of results of MPSA procedure are plots of empirical cumulative distribution functions (ECDF) of acceptable and unacceptable samples, for each of the selected parameters. The final result is <paramRankingXmlList>. It contains two rankings of parameters. Values of rankings are calculated by comparing ECDFs with Kolmogorov-Smirnov test (KS test) and one minus Pearson product-moment correlation coefficient (PMCC). Below are presented results for simple enzymatic reaction.
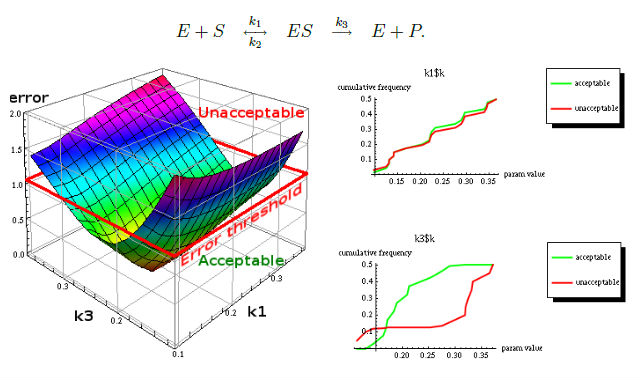
More experiments are available on website http://bioputer.mimuw.edu.pl/tav4sb/.
Links:
Publications :
- Rybiński, M. et al., 2012, Tav4SB: integrating tools for analysis of kinetic models of biological systems, BMC Systems Biology, 6(1), p.25.
- Rybiński, M. et al., 2011, Tav4SB: grid environment for analysis of kinetic models of biological systems, ISBRA 2011 Short Abstracts, pp. 92-95.

 Polski
Polski English
English