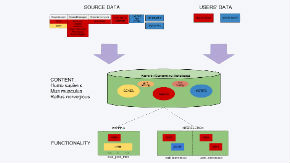
 Nencki Genomics Database (NGD) contains data on regulatory regions on the scale of the whole genomes of human, mouse and rat. Our database is the first regulatory regions database for the rat. NGD is based on the reference Ensembl funcgen database, the content and functionality of which it extends with:
Nencki Genomics Database (NGD) contains data on regulatory regions on the scale of the whole genomes of human, mouse and rat. Our database is the first regulatory regions database for the rat. NGD is based on the reference Ensembl funcgen database, the content and functionality of which it extends with:
- ability to add users’ data, with access control of the added data by the users;
- results of motif finding for known transcription factor binding motifs (Jaspar, Transfac Professional) on the scale of whole genomes, and the results of finding of evolutionally conserved non-coding sequences;
- stored procedures for computing intersections, between regulatory regions, and between regulatory regions and instances of transcription factor binding motifs, as well as procedures to map regulatory areas to the genes.
The database content and the stored procedures are accessible from a mysql client and through the layer of webservices (recommended).
Access from a web browser is possible via the Nencki Genomics Portal, and permits visualization of the database content for the user-selected genes, and export of these data to files of tabular format. Publicly accessible data added by the users can also be visualized in the Ensemble Genome Browser, as a DAS track.
Links:
- http://galaxy.nencki-genomics.org (dostęp z poziomu przeglądarki)
- http://www.nencki-genomics.org (dokumentacja i rejestracja)
References:
- Krystkowiak I, Lenart J, Debski K, Kuterba P, Petas M, Kaminska B, Dabrowski M. Nencki Genomics Database–Ensembl funcgen enhanced with intersections, user data and genome-wide TFBS motifs. Database (Oxford). 2013 Oct 1;2013:bat069. doi: 10.1093/database/bat069.

 Polski
Polski English
English